
As we cut both the DNA fragment containing our gene and the plasmid with the same enzyme, sticky ends form in each of them. Let’s say we would like to insert our gene of interest into a vector plasmid.

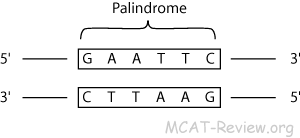
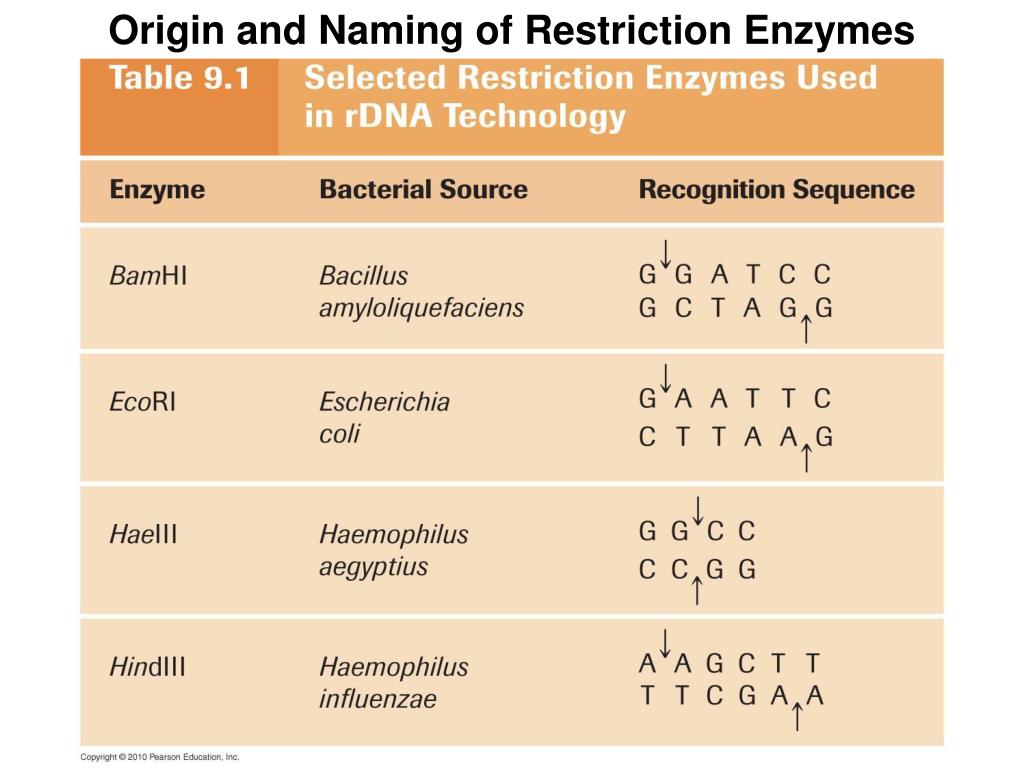
Because these overhangs are capable of annealing with complementary overhangs, they are called “sticky ends.” Sticky ends facilitate the production of recombinant DNA. Many restriction endonucleases make staggered cuts in the two strands of DNA, such that the cut ends have a short single-stranded overhang. Therefore, its digestion with a restriction enzyme generates many smaller DNA fragments – restriction fragments. As one would expect, a long DNA molecule naturally has many such restriction sites. As soon as this recognition occurs, enzyme cuts the sugar-phosphate backbone from specific points, which are generally within the restriction site. For example, the sequence ATTGCAAT is palindromic. Most restriction enzymes recognize palindromic sequences, meaning that both strands of DNA will have the same sequence when read 5′ to 3′. This is a short DNA sequence (generally 4-8 nucleotide pairs) that an enzyme specifically recognizes. What makes restriction enzymes suitable for gene cloning is that each of them has a specific restriction site. As they are endonucleases, they can cut foreign DNA from the inside and make it ineffective. Naturally, they are defense systems of bacteria against foreign DNA. restriction endonucleases) are part of the genetic engineering toolbox and make gene cloning possible. So I think the last sentence is looking at things from the wrong perspective.Restriction enzymes recognize specific DNA sequences and cut them in a predictable manner. There is no particular “advantage for bacteria” other than “it works”. There is also a PDB Molecule of the Month item on this, although unfortunately the bending of the DNA is not apparent in the images there.Ĭomment on Last Two Sentences of Question
This answer is based on chapter sections 9.3.2 and 9.3.3 of Berg et al., which is available online at NCBI Bookshelf and is well worth reading in full. DNA lacking the recognition site is not cleaved by the restriction enzyme because it does not bind specifically to the protein so that the magnesium ion remains too far away. Methylation of the adenine nucleotide in the recognition sequence prevents hydrogen bonding of the enzyme to the DNA. Compare bending a thin pipe over one’s knee, applying equal force with both hands.) (The dimeric nature of enzyme allows force to be exerted at two points, to produce the bend.

This binding causes the DNA to bend (C) bringing a catalytic magnesium ion into a position in which it can interact with phosphate of the phosphodiester bond that is being cleaved (D). The ‘palindromic’ nature of the recognition/cleavage site of restriction endonucleases, such as EcoRV (illustrated here) results in a form of symmetry (A) that allows a dimeric enzyme - that forms a tunnel - to bind symmetrically at that site in the DNA (B). Way that facilitates the endonuclease reaction. The palindromic symmetry of the restriction site allows a dimericĮnzyme to bind the DNA in a manner that bends the double helix in a


 0 kommentar(er)
0 kommentar(er)
